Week 3 -- January 23rd, 2015
23 Jan 2015The next meeting for the class is on Friday at 12:50pm, in Carr 611 as always.
We will start with the project pitches, and we will continue with another
lecture that will cover one of the most important data structure in R, the
data.frame.
The project pitches this week is a first iteration (we’ll do it again next week) that will help articulate what you will be working on this semester.
The project should be useful for you and/or your research. You should be excited about it so you are motivated to spend time working on it, and going through the (sometimes frustrating) process that learning a new programming language can be.
If possible, I would like your project to be divided into 3 goals that can be completed in steps. The first step should be relatively easy to complete, and the next 2 steps a little harder as the previous ones. Without programming experience, I realize that it might be difficult for you to identify how hard a given task is. Even with some experience, I always underestimate how long and how difficult something takes to program. Another question you might have is, “I am not even sure what I can do with R?” in other words “you don’t know what you don’t know”. For the most part, I know as much as you when it comes to analyzing genomic or spatial data in R. So, we’ll have to figure out together the details of your project.
I am organizing these project pitches for several reasons, one being that I want to give you the opportunity to hear about other people projects so you might feel excited about it, and collaborate with them. Another is that it will help you identifying people who will likely face similar challenges.
The pitches should last a total of 5 minutes (3 minutes for the pitch and 2 minutes for questions/discussion). Slides are not required. If you do use slides, I strongly encourage you to use an online platform (Google Slides, upload your presentation to slideshare, etc…) so we don’t spend too much time switching computers on the projector.
In addition of your presentations, I would like you to write a couple of paragraphs about your projects. I will use these notes to provide feedback for next week.
Your presentation and your notes should include enough information for me to be able to evaluate the feasibility of your projects. These notes should include:
- the general goal of your project
- the hypotheses you would like to test
- the kind of data you would like to analyze (file format, whether the data is ready to be analyzed or need to be cleaned up beforehand…)
- the type of analysis you would like to carry
- the R packages you know will be useful for your project
Your projects and your presentation can be done on your own or in groups of 2-3.
In summary, for Friday, I encourage you to:
- Review the content of last week lecture
- Read the “How to get help?”
- Prepare the notes associated with your presentation (Instructions below)
EDIT: The notes from the lecture are now available
How to prepare the notes for your presentation?
- Go to https://github.com/r-bio/logistics
- Click on the “+” next to “logistics” as image below to create a new file.
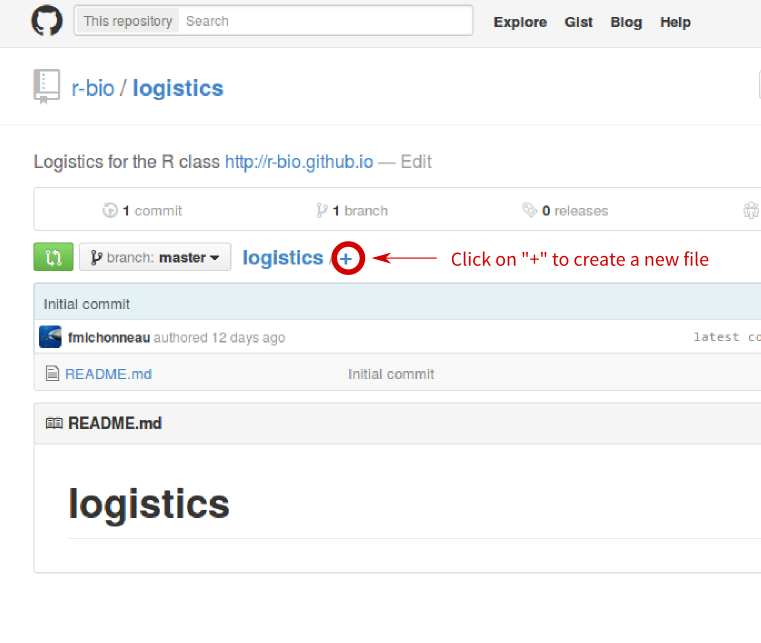
- Name your file
Lastname_Firstname.md(e.g., mine would have beenMichonneau_Francois.md). Enter your text in the box below. Once you are done, click on the green button at the bottom of the page.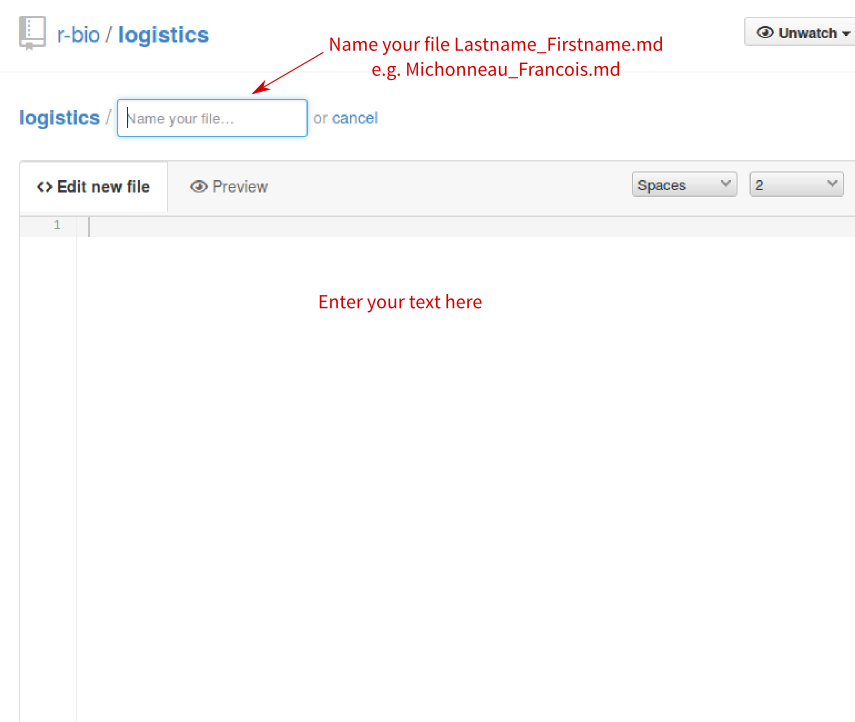
If you need to go back to your page to edit it:
- go to https://github.com/r-bio/logistics
- click on your file name
- click on the pencil icon on the top right corner.
- once you are done with your edits, click on the green button at the bottom of the page.
.md stands for “Markdown”. It is a popular and lightweight way of converting a
plain text file into something pretty. You don’t have to worry about any kind of
formatting for the notes accompanying your presentation, but if you want to
explore and learn more about it, check out
this page.